Download Software
Currently supported Software:
 |
Go to the FiberViewerLight webpage | Fiber ViewerLight is an open-source C++ application to analyze fiber bundles. It provides: |
 |
Go to the DTIAtlasFiberAnalyzer (DTI Fiber Tract Statistics) webpage | DTIAtlasFiberAnalyzer is an open-source C++ application that applies an atlas fiber to all individual subjects from a dataset, extracts fiber profiles, gathers and plots related information (FA map…)
|
| DTI-Reg | Go to the DTI-Reg webpage | DTI-Reg is an open-source C++ application that performs pair-wise DTI registration, using scalar FA map to drive the registration.
|
| Tractograpy with unscented Kalman Filter | Go to the UKF webpage | We present a framework which uses an unscented Kalman filter to perform tractography. At each point on the fiber the most consistent direction is found as a mixture of previous estimates and of the local model. Currently it is possible to tract fibers using two different 1-, 2-, or 3-tensor methods. |
| Go to the DTIAtlasBuilder webpage | This tool creates an Atlas image as an average of several DTI images that will be registered. The registration will be done in two steps : A final step will apply the transformations to the original DTI images so that the final average can be computed. |
|
| DTI Process Toolkit | Go to the DTI Process webpage | DTIProcess is a DTI processing and analysis toolkit developed in UNC and University of Utah. Tools in this toolkit include dtiestim, dtiprocess, dtiaverage, fibertrack, fiberprocess…
|
|
|
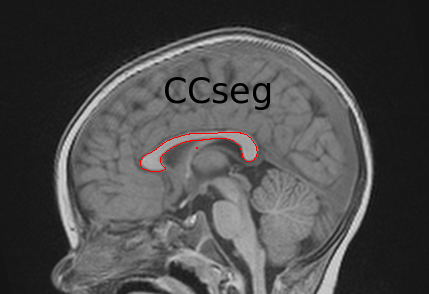
Go to the CCSeg webpage | CCSeg is an open-source C++-based application that allows automatic as well as user-interactive segmentation of the Corpus Callosum.
Via a Qt-based graphical user interface, CCSeg also performs semi-automatic segmentation. |
|
|
Go to the NIRAL Utilities webpage | NIRAL Utilities are open-source C++ based command line applications that allow image analysis and processing using ITK or VTK libraries |
 |
Go to the ShapeWorks webpage | ShapeWorks software is an open-source distribution (initially developped by the SCI Institute) of a new method for constructing compact statistical point-based models of ensembles of similar shapes that does not rely on any specific surface parameterization: |
|
|
Go to the DTIPrep webpage | DTIPrep performs a “Study-specific Protocol” based automatic pipeline for DWI/DTI quality control and preparation: |
|
|
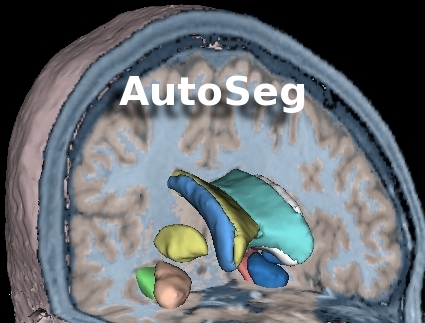
Go to the AutoSeg webpage | AutoSeg is an end-to-end application that performs automatic brain tissue classification and structural segmentation: |
|
|
Go to the GAMBIT webpage | GAMBIT is an end-to-end application allowing Group-wise Automatic Mesh-Based analysis of cortIcal Thickness as well as other surface area measurements: |
|
|
Go to the ARCTIC webpage | ARCTIC (Automatic Regional Cortical ThICkness) is a cross-platform end-to-end application allowing individual analysis of cortical thickness. It can be run within Slicer3 as an external module or directly as a command line: |
|
|
Go to the NITRC SPHARM-PDM page | The SPHARM shape analysis toolset is used for doing local statistical shape analysis. ITK based. |
|
|
Go to the NITRC MriWatcher page | MriWatcher is a vizualisation tool for MRI images (.gipl,.mha,.hdr). |
|
|
Go to the HeadCircumference webpage | Program which compute head circumference based on a 3D image |
|
|
Go to the MakeAtlasScripts – Rodent webpage | Sets of BatchMake scripts to process rodent images
|
Non-supported Software:
NeuroLib download page
CCSeg is an open-source C++-based application developed at UNC-Chapel Hill that allows automatic as well as user-interactive segmentation of the Corpus Callosum. Via a Qt-based graphical user interface, CCSeg also performs semi-automatic segmentation